I’ve recently been communicating with Ebrahim Afshinnekoo who is Project Director for the PathoMap project based at the Weill Cornell Medicine Mason Laboratory in New York City. Launched in the summer 2013, PathoMap was the first project of its kind, with the intent to comprehensively map and investigate the presence of bacteria and DNA on the surfaces of large urban, metropolitan environments such as New York City. And of course what better venue to collect bacteria samples in NYC than the subway system – the large subterranean behemoth home to 5.5 million riders on an average weekday.
I was drawn to the project in that it involves several common geospatial components the traditional GIS community is routinely involved with such as data collection/data validation, data analysis, mobile apps, web mapping and visualization. To date, discussion on this geospatial research effort has focused mainly within the Cell Systems (scholarly journal) community, though with little exposure within the traditional NYS GIS community. While both the Wall Street Journal and the New York Times published articles on PathoMap in 2015 we’ve seen little work of this nature at statewide conferences or how it can promote similar geospatial analysis across the Empire State. With this in mind, eSpatiallyNewYork initiated this blog entry with the purpose of exposing the PathoMap project, and its subsequent global expansion (MetaSUB) to the larger statewide GIS community.
Data Collection
The molecular profiling initiative launched in the summer of 2013 with the help of undergraduates from Cornell University and Macaulay Honors College – which were soon to be given the appropriate moniker “Swab Squad”. To create a city-wide profile, the research team first built an Android/iOS mobile application in collaboration with GIS Cloud to enable real-time entry and loading of sample metadata directly into a database (Figure 1).
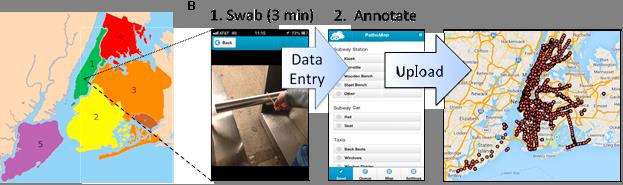
Figure 1: Data collection from the project included the “swabbing” of sites and subsequent analysis and data entry of the findings into a mobile app which are dynamically uploaded to the Cloud GIS database. Source: Afshinnekoo et al., 2015
